Firewalled Computing Resources Overview
Doing Work and Computing
When doing research, running jobs and the like, please be careful of your resource consumption on the server you are on. Don't run too many threads or cores at once if such a thing overruns the RAM available or the disk IO available. If you are not sure of your potential RAM, CPU or disk impact, start small with one or two processes and work your way up from there. Also, before running your stuff, check what else is already happening on the server by using the 'top' command to see who else and what else is running and what kind of resources are already being consumed. If, after starting a process, you realize that the server slows down considerably or becomes unusable, kill your processes and re-evaluate what you need to make things work. These servers are shared resources - be a good neighbor!
Server Types and Management
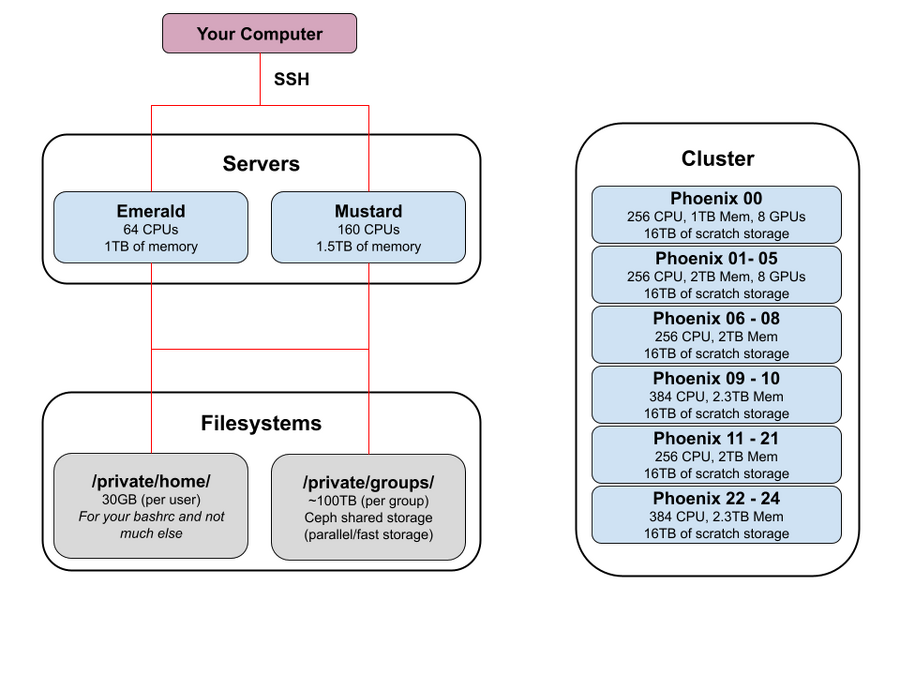
After confirming your VPN software is working, you can ssh into one of the shared compute servers behind the VPN. The DNS suffix for all machines is ".prism". So, "mustard" would have a full DNS name of "mustard.prism":
| Node Name | Operating System |
CPU Cores | Memory | Network Bandwidth | Scratch Space |
|---|---|---|---|---|---|
| mustard | Ubuntu 22.04 | 160 | 1.5 TB | 10 Gb/s | 9 TB |
| emerald | Ubuntu 22.04 | 64 | 1 TB | 10 Gb/s | 690 GB |
| crimson | Ubuntu 22.04 | 32 | 256 GB | 10 Gb/s | 5.5 TB |
| razzmatazz | Ubuntu 22.04 | 32 | 256 GB | 10 Gb/s | 5.5 TB |
These shared servers are managed by the Genomics Institute Cluster Admin group. If you need software installed on any of these servers, please make your request by emailing cluster-admin@soe.ucsc.edu. The above mentioned scratch space is usually located in /data/scratch on each server.
The Firewall
All servers are behind a firewall in this environment, and as such, you must connect to the VPN in order to access them. They will not be accessible from the greater Internet without VPN. Although you will be able to connect outbound from them to other servers on the internet to copy data in, sync git repos, stuff like that. It is only inbound connections that will be blocked. All machines behind the firewall have the private domain name suffix of "*.prism".
The Phoenix Cluster
This is a cluster of 25 Ubuntu 22.04 nodes, some of which have GPUs in them. Each node generally has about 2TB RAM and 256 cores, although the cluster is heterogeneous and has multiple node types. You interact with the Phoenix Cluster via the Slurm Job Scheduler. You must specifically request access to use Slurm on the Phoenix Cluster, just email cluster-admin@soe.ucsc.edu for access.
| Node Name | Operating System |
CPU Cores | GPUs/Type | Memory | Network Bandwidth | Scratch Space |
|---|---|---|---|---|---|---|
| phoenix-00 | Ubuntu 22.04 | 256 | 8 / Nvidia A100 | 1 TB | 10 Gb/s | 16 TB NVMe |
| phoenix-[01-05] | Ubuntu 22.04 | 256 | 8 / Nvidia A5500 | 2 TB | 10 Gb/s | 16 TB NVMe |
| phoenix-[06-08] | Ubuntu 22.04 | 256 | N/A | 2 TB | 10 Gb/s | 16 TB NVMe |
| phoenix-[09-10] | Ubuntu 22.04 | 384 | N/A | 2.3 TB | 10 Gb/s | 16 TB NVMe |
| phoenix-[11-21] | Ubuntu 22.04 | 256 | N/A | 2 TB | 10 Gb/s | 16 TB NVMe |
| phoenix-[22-24] | Ubuntu 22.04 | 384 | N/A | 2.3 TB | 10 Gb/s | 16 TB NVMe |
The cluster head node is phoenix.prism. However, you cannot directly login to phoenix.prism in order to protect the scheduler from errant or runaway jobs there, so jobs can be submitted from any interactive compute server (mustard, emerald, razzmatazz or crimson). To learn more about how to use Slurm, refer to:
https://giwiki.gi.ucsc.edu/index.php/Genomics_Institute_Computing_Information#Slurm_at_the_Genomics_Institute
For scratch on the cluster, TMPDIR will be set to /data/tmp (which is local to each cluster node). That area is cleaned often so don't store any data there that isn't being used by your jobs.
Graphical Diagram of the Firewalled Area
This is a general representation of how things look: